RNA Genomics Laboratory

Research Topics
I. Regulation of RNA stability by post-transcriptional gene regulation
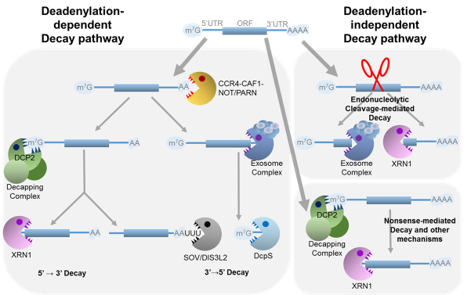
The stability of mRNAs is regulated by several pathways, including deadenylation-dependent degradation, decapping, and endonucleolytic cleavage. We study these pathways by profiling degraded mRNAs and identifying various mRNA features that influence stability.
II. Small RNA-mediated gene expression regulation
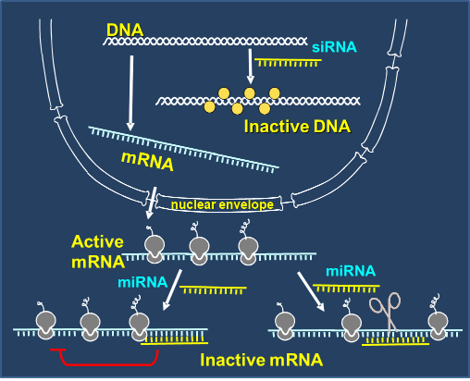
Small RNAs are noncoding RNAs that regulate gene expression via transcriptional and post-transcriptional regulation. We mainly study the role of miRNA and siRNAs in developmental process and environmental stress response of plants.
III. Plant development and environmental stress resistance
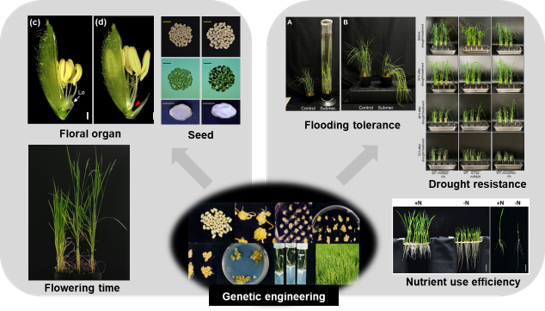
Genetic engineering through CRISPR-mediated genome editing and transformation enables us to study genes involved in plant development and environmental stress resistance.
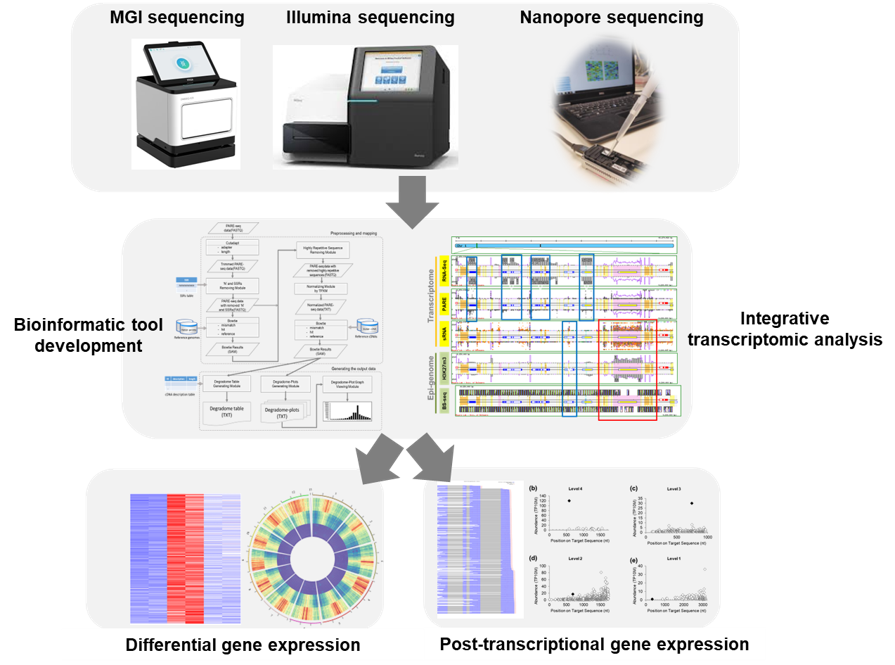
Research Tools
Lab Members
Current members
-
-

-
Dong-Hoon Jeong
(Principle Investigator)
-
-
-

-
So Young Park
(Postdoctoral Researcher)
-
-
-

-
Thi Tuyet Suong Ha
(Postdoctoral Researcher)
-
-
-

-
Ah Hyeon Lee
(Master Student)
-

Former members
- Hyo Jin Heo (Master student)
- Hyeon Cheol Park (Master student)
- Min Seo Hwang (Master student)
- Sang Yoon Lee (Master student)
Research Publications
Selected publications
- 1.SY Park, HS Chang, SH Lim, D-H Jeong, YJ Kim (2024) Functional characterization of PRP43a, a component for splicing disassembly in Arabidopsis. Journal of Plant Biology 67 (6), 427-435
- 2.SY Park, D-H Jeong (2023) Comprehensive analysis of rice seedling transcriptome during dehydration and rehydration. International Journal of Molecular Sciences 24 (9), 8439
- 3.SY Park, J Cho, D-H Jeong (2022) Small regulatory RNAs in rice epigenetic regulation. Biochemical Society Transactions 50 (3), 1215-1225
- 4.JM Shin, J Jeon, D Jung, K Kim, YJ Kim, D-H Jeong, JH Yoon (2022) PhenGenVar: a user-friendly genetic variant detection and visualization tool for precision medicine. Journal of Personalized Medicine 12 (6), 959
- 5.JI Won, JM Shin, SY Park, JH Yoon, D-H Jeong (2020) Global analysis of the human rna degradome reveals widespread decapped and endonucleolytic cleaved transcripts. International journal of molecular sciences 21 (18), 6452
- 6.JI Won, JB Lee, HW Lee, JM Shin, JH Yoon, DH Jeong (2019) WebPORD: A web-based pipeline of RNA degradome. International Journal of Data Mining and Bioinformatics 22 (3), 216-230
- 7.Jeong D-H, Schmidt SA, Rymarquis LA, Park S, Ganssmann M, German MA, Accerbi M, Zhai J, De Paoli E, Fahlgren N, Fox SE, Garvin DF, Mockler TC, Carrington JC, Meyers BC, and Green PJ. (2013) Parallel analysis of RNA ends enhances global investigation of miRNAs and target RNAs of Brachypodium distachyon. Genome Biology 14:R145
- 8.Jeong D-H, Park S, Zhai J, Gurazada SGR, De Paoli E, Meyers BC, and Green PJ. (2011) Massive analysis of rice small RNAs: Mechanistic implication of regulated miRNAs and variants for differential target RNA cleavage. Plant Cell 23: 4185-4207
- 9.Jeong D-H, An S, Park S, Kang H-G, Park G-G, Kim S-R, Sim J, Kim Y-O, Kim M-K, Kim S-R, Kim J, Shin M, Jung M, An G. (2006) Generation of flanking sequence tag database for activation tagging lines in japonica rice, Plant J. 45:123-132.
- 10.Jeong D-H, An S, Kang H-G, Moon S, Han J-J, Park S, Lee H S, An K, An G. (2002) T-DNA insertional mutagenesis for activation tagging in rice. Plant Physiol. 130:1636-1644.


